Masood Ayoub* , Bilal Ahmad Bhat1, Shahjahan Ul Islam1,2, Syed Masood Ahmad Rizvi2 and Qazi Mohd Junaid3
, Bilal Ahmad Bhat1, Shahjahan Ul Islam1,2, Syed Masood Ahmad Rizvi2 and Qazi Mohd Junaid3
1Department of Chemistry, Govt. Degree College Shopian, Jammu and Kashmir-192303
2Department of Chemistry, University of Kashmir, Hazratbal Srinagar, Jammu and Kashmir-190006
3Department of Chemistry, Pondicherry University, Pondicherry-605014
Corresponding Author Email : makandchem@gmail.com, masoodku2@gmail.com
DOI : http://dx.doi.org/10.13005/msri/160306
Article Publishing History
Article Received on : 22-Oct-2019
Article Accepted on : 14-Dec-2019
Article Published : 17 Dec 2019
Plagiarism Check: Yes
Reviewed by: Mehdihasan Shekh Second Review by: Mirestean Camil Ciprian
Final Approval by:
Manoj Gupta
Second Review by: Mirestean Camil Ciprian
Final Approval by:
Manoj Gupta
Article Metrics
ABSTRACT:
The design and development of synthetic fluorescent molecular architectures for sensing of nucleic acids and related species in living cells is an area of enormous interest. For the first time a novel compilation of single molecular abiotic fluorescent receptors for nucleic acid detection in living cells have been reviewed. Selected reports have been screened and thoroughly discussed which have revealed enormous promise for bio imaging. The mechanistic aspects of nucleic acid, phosphate or nitrogenous base sensing upon encounter with the receptors has been examined under diverse matrices. In addition to the cytotoxicity, specific conditions deciphering suitable and promising results for real-time application have been highlighted.
KEYWORDS:
Cytosine; Fluorescence; DNA; Nucleic Acid; Phosphate; Receptor
Copy the following to cite this article:
Kaloo M.A, Bhat B.A, Islam S, Rizvi S.M.A and Junaid Q.M. Abiotic Fluorescent Receptors for Bioimaging: Sensing of Nucleic Acids.Mat. Sci. Res. India; 16 (3).
|
Copy the following to cite this URL:
Kaloo M.A, Bhat B.A, Islam S, Rizvi S.M.A and Junaid Q.M. Abiotic Fluorescent Receptors for Bioimaging: Sensing of Nucleic Acids.Mat. Sci. Res. India; 16 (3). Available from: http://bit.ly/2svZKAX
|
Introduction
At present, the concept of molecular recognition and its utility for living system applications is fundamental from both chemistry and biomedical point of view1-3. The interaction of synthetic molecules “receptors” with nucleic acid (DNA) moiety and related species inside living cells is highly fruitful, and an area of great interest particularly in biomedical research, diagnostic and cure of deadly diseases.4-6 With such significant considerations, a number of abiotic single molecular receptors and their interaction with the nucleic acid moiety and allied cellular fragments has been studied in detail.7-8 The encounter between receptor and nucleic acid fragments has been recognized through efficient signal transduction in the form of colour and fluorescence. Among various strategies, it is the fluoremetric based receptor approach which decodes real-time applications owing to enormous sensitivity and visual signal readout of events under complex biological environments.9-13 Keeping all these considerations in mind, in this review we have discussed only some specific and selected works of chemical sensing which are either directly related to DNA, nitrogenous base, phosphate group, etc.
Receptors for DNA sensing
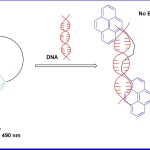
In this direction, C Schmuck et al (2012)14 reported a DNA sensing receptor 1 (Fig 1) which was evaluated with HeLa cell lines for DNA imaging. According to their observation, at 4 °C, HeLa cells could not uptake receptor molecule and hence no DNA staining was achieved. However, at 37 °C uptake of receptor into the cells was obtained, and was followed by subsequent staining of nuclear DNA. It has been observed that 1 does not possess any general cytotoxicity to living cells and hence was utilized as a tool of potential interest for diagnostic applications. It has been distinguishly observed that upon binding of 1 to HeLa cells, conformation of 1 changes from folded to extended form, which leads to a distinct change in the fluorescence characteristics of the molecule. Thus it can be used for imaging of nucleic acid in living cells. Once the interaction was studied through emission spectroscopy, 1 shows strong emission at λ = 490 nm and a weaker one at λ = 406 nm. These bands were attributed to a single pyrene chromosphere and a pyrene excimer respectively. However, upon binding to DNA, the monomer emission increased while excimer emission decreased accordingly and finally resulting in complete disappearance.
Figure 1. Mechanism of receptor 1 and DNA interaction.
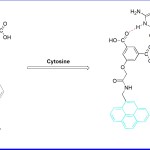
In another study, P Sahoo et al (2017)15 reported a new and a simple receptor 2 (Fig 2) which showed efficient fluorescence readout upon selective binding with free cytosine under aqueous conditions in living cells. Before interaction with the cytosine moiety, fluorescence of 2 was found quenched due to benzoic acid moiety. However after interaction, the fluorescent property was recovered. Thus via this work, one could detect free cytosine in human cancer cells where its concentration always remains very high. Once the mechanistic aspects were examine under emission spectroscopy, a remarkable enhancement in fluorescence intensity at 377 nm and 395 nm was obtained upon binding of cytosine with receptor. The binding constant (Ka) for 2 with cytosine was calculated as 3.48 × 105 M-1. The limit of detection was calculated to be around 32 nM at neutral pH. The “turn on” fluorescence of receptor 2 in the presence of free cytosine was attributed to the intermolecular hydrogen bonding with cytosine. This in turn promotes blockade of photoinduced electron transfer (PET) in the receptor.
Figure 2. Mechanism of receptor 2 and Cytosine interaction.
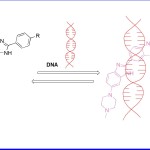
In the similar direction, J Bucevicius et al (2018)16 reported a series of Hoechst dye 3 (Fig c) for the detection of nucleic acid (DNA) in living cells, tissues, animals and fixed cells. These receptors were found specifically staining the nucleus and were reported to be less cytotoxic. The dyes were also observed to have high affinity and specificity towards DNA. This property attributes them as excellent target moieties for nucleic acid detection in living systems. Binding of Hoechst moiety to DNA supresses this interaction and increases fluorescent intensity. Once examined under emission spectroscopy, a remarkable enhancement in fluorescence intensity at 358 nm along with its shift towards 460 nm was obtained upon binding of receptor with DNA.
Figure 3. Mechanism of receptor 3 (& derivatives) and DNA interaction.
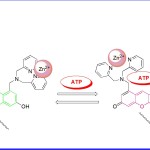
I Hamachi et al (2012)17 reported a novel fluorescent receptor 4 (Fig 4) in order to monitor different cell imaging process in living cells. He designed an auto localizable fluorescent receptor to detect the release of NPPs by the cell to its exterior as well as the stimuli-responsive change in ATP concentration inside mitochondria. It was found that the receptor complex with Zn (ΙΙ) showed high affinity for ATP and the changes were observed via fluorescent signal. This sensing of ATP by Zn (ΙΙ) receptor complex involved the binding-induced recovery of a fluorescent conjugated form of a xanthene ring from its non fluorescent deconjugated form. In order to achieve the detection and imaging of NPPs on the plasma membrane surface, a lipid anchor was introduced into the xanthenes-based Zn (II) receptor complex. Spectroscopic studies revealed that the developed receptor could sense NPPs with several fold increase of their fluorescence intensities through a sensing mechanism similar to earlier. In the live cell application, receptor Zn (II) complex containing a lipid anchor localized on the plasma membrane surface and could detect the extracellular release of NPPs during cell necrosis induced by streptolysin. Once the discussed interactions between various target species and receptor/receptor (Zn2+) complexes were examined under fluorescence spectroscopy, strong enhancement in the fluorescent intensity was revealed.
Figure 4. Mechanism of receptor 4 and ATP interaction
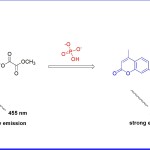
In another important study, J Yoon et al (2014)18 reported a receptor 5 (Fig 5) which selectively detects endogenous inorganic phosphate (pi) in hemi channel-closed cells. In this case, the receptor interacts with inorganic phosphate and undergoes hydrolytic reaction in HEPES-DMSO (V/V=1:9) buffered (0.02 M, pH 7.4). The phosphate induced hydrolytic reaction results in the 780-fold enhancement of fluorescent intensity of peak around 455 nm. It is believed that upon interaction of receptor with pi, coumarin formation takes place which deciphers the emission signal around 455 nm. The mechanism was fully supported by spectroscopy and theoretical calculations. The obtained behavior of the receptor was achieved with HeLa cells. Receptor 5 was also utilized to elucidate the mechanism of Inx3 action in hemichannel-closed Sf9 cells.
Figure 5. Mechanism of receptor 5 and ATP interaction.
Conclusions
In this work, herein we have presented and highlighted important and significant efforts put forward by various scientists and bio engineers across the globe in the field of nucleic acid imaging. We have worked on certain crucial reports which deliver and decipher enormous promise for real-time applications. Among these, mechanistic aspects of nucleic acid, phosphate or nitrogenous base interaction with the fluorescent receptor have been presented. Even though efforts made by researchers are worth appreciating, but none the less there are still enough insights to look into, particularly the complexity in the design of the single molecular receptor platform. In addition, aspects like use of toxic metal ions for design of metal-complex based sensors, chemical species released as a result of some hydrolytic reaction, etc and their toxic implications need to be worked out. Most importantly, design of reversible receptor frameworks to target diverse species inside body and cells is highly demanding. We believe the current-state of art of receptors mentioned in this work will inspire the researchers interested in the field and allied, and will provide them a quick platform for future endeavour.
Acknowledgments
M. A. Kaloo gratefully acknowledges the rtment of chemistry at Govt. Model Degree College (GMDC) Shopian for support and DST, New Delhi for Faculty Award (DST/INSPIRE/04/2016/000098).
Funding Source
This research received no specific grant from any funding agency.
Conflict of Interest
The authors have no conflicts of interest to disclose.
References
1. S. Dey, D. Saina and S. P. Goswami, RSC Adv., 2014, 4, 428–433.
2. M. C. Heffern, H. M. Park, H. Y. Au-Yeung, G.C. Van de Bittner, C. M. Ackerman, A. Stahl and C. J. Chang, Proc. Natl. Acad. Sci., 2016, 113, 14219–14224.
3. D. Wu, A. C. Sedgwick, T. Gunnlaugsson, E. U. Akkaya, J. Yoon and T. D. James, Chem. Soc. Rev., 2017, 46, 7105–7123.
4. P. Sahoo, Bioorg. Chem., 2015, 58, 26–47.
5. B. Jiang, B, G. H. Tang, K. Cao, L. Y. Wu, R. Wang, Antioxid. Redox Signaling 2010, 12, 1167−1178.
6. Z. Guo, G. Chen, G. Zeng, Z. Lia, A. Chen, J. Wang, L. Jiang, 2014, DOI: 10.1039/C4AN01909A. All need to inreoduced except 9…..
7. P. Kamoun, M. C. Belardinelli, A. Chabli, K. Lallouchi, B. Chadefaux-Vekemans, Am J Med Genet A. 2003, 116A, 310−311.
8. M. M Gadalla, S. H. J. Snyder, Neurochem. 2010, 113, 14-26.
9. W. Saenger, Principles of Nucleic Acid Structure, Springer-Verlag, New York, 1988.
10. S. Erten-Ela, M. D. Yilmaz, B. Lcli, Y. Dede, S. Lcli and E. U. Akkaya, Org. Lett. 2008, 10, 3299.
11. R. Guliyev, A. Coskun and E. U. Akkaya, J. Am. Chem. Soc. 2009, 131, 9007.
12. L. -Y. Niu, Y. -S. Guan, Y. -Z. Chen, L. -Z. Wu, C. -H. Tung and Q. -Z. Yang, J. Am. Chem. Soc. 2012, 134, 18928.
13. A. Loudet and K. Burgess, Chem. Rev. 2007, 107, 4891.
14. J. Wu, Y. Zou, C. Li, W. Sicking, I. Piantanida, T. Yi and C. Schmuck, J. Am. Chem. Soc. 2012, 134, 1958−1961.
15. H. S. Sarkar, S. Das, D. Mandal, M. R. Uddin, S. Mandal and P. Sahoo, RSC Adv., 2017, 7, 54008-54012.
16. J. Bucevicius, G. Lukinavicius, and R. Gerasimait, Chemosensors 2018, 6, 18-30.
17. Y. Kurishita,T, T. Kohira, A. Ojida, and I. Hamachi, J. Am. Chem. Soc., 2012, 134, 18779−18789.
18. L Guo, J F Zhang,Xin Yi Liu, Li Mei Zhang, Hong Li Zhang,J H Chen, X G Xie,Ying Zhou, K Luo and J Yoon, Anal Chem., 2014, 1-8.

This work is licensed under a Creative Commons Attribution 4.0 International License.
 , Bilal Ahmad Bhat1, Shahjahan Ul Islam1,2, Syed Masood Ahmad Rizvi2 and Qazi Mohd Junaid3
, Bilal Ahmad Bhat1, Shahjahan Ul Islam1,2, Syed Masood Ahmad Rizvi2 and Qazi Mohd Junaid3
 Material Science Research India An International Peer Reviewed Research Journal
Material Science Research India An International Peer Reviewed Research Journal